
In February 2024, a study led by Professor Hongyan Tong from the First Affiliated Hospital, Zhejiang University School of Medicine was published in the international academic journal ——Journal of Clinical Investigation(IF=15.9). The title of the study is “Ectopic expression of transcription factor ONECUT3 drives complex karyotype in Myelodysplastic Syndromes“. This study provides compelling evidence for the critical role of the transcription factor ONECUT3 in driving complex karyotypes and poor outcomes in Myelodysplastic Syndromes (MDS).
This study investigates the role of the transcription factor ONECUT3 in Myelodysplastic Syndromes (MDS) characterized by complex karyotypes (CK). The research identifies a correlation between high ONECUT3 expression and the presence of specific genetic mutations, such as PHF6 and wild-type IDH2 and KMT2D, suggesting an oncogenic role for ONECUT3 in MDS pathophysiology.
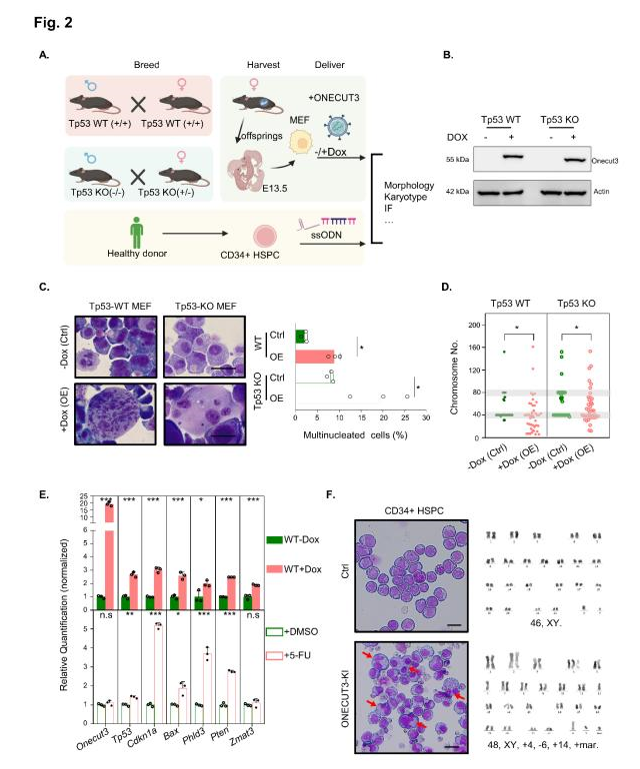
Myelodysplastic Syndromes (MDS) represent a heterogeneous group of clonal hematopoietic disorders characterized by ineffective hematopoiesis and morphological dysplasia. One of the significant challenges in managing MDS is the identification of factors contributing to disease progression, particularly in cases with complex karyotypes (CK). Complex karyotypes are associated with poor prognosis and increased risk of transformation to acute myeloid leukemia (AML). While mutations in TP53 are well-established drivers of CK in MDS, recent research has shed light on the role of transcription factors like ONECUT3 in influencing karyotypic complexity, independently of TP53 mutations. This study delves into the mechanisms by which ONECUT3 contributes to chromosomal instability and poor outcomes in MDS patients with CK, presenting implications for targeted therapeutic interventions.
To comprehensively investigate the role of ONECUT3 in MDS with complex karyotypes, a rigorous study design was implemented, incorporating various experimental techniques and analytical approaches. Initially, patient samples diagnosed with MDS and exhibiting complex karyotypes were collected, ensuring representation across different genetic backgrounds and disease subtypes. RNA sequencing analysis was performed on these samples to quantify ONECUT3 expression levels and correlate them with specific genetic mutations commonly associated with MDS, including PHF6, IDH2, and KMT2D.
Furthermore, chromatin immunoprecipitation sequencing (ChIP-seq) was employed to identify genomic regions bound by ONECUT3, providing insights into its transcriptional targets and potential downstream effects. By characterizing the binding profile of ONECUT3 in MDS cells, this approach aimed to elucidate the molecular pathways through which ONECUT3 contributes to chromosomal instability and disease progression.
In parallel, cell culture models of MDS were utilized to assess the functional consequences of ONECUT3 dysregulation. Cell viability assays were conducted to evaluate the effects of modulating ONECUT3 expression levels on cell proliferation and survival. Additionally, quantitative analysis of gene expression levels was performed to elucidate the downstream signaling pathways activated by ONECUT3 in MDS cells, shedding light on its mechanistic role in driving complex karyotypes.
The comprehensive analysis of patient samples revealed a significant correlation between elevated ONECUT3 expression levels and the presence of specific genetic mutations, including PHF6, wild-type IDH2, and wild-type KMT2D, in MDS patients with complex karyotypes. These findings underscored the oncogenic potential of ONECUT3 in driving chromosomal instability and disease progression independently of TP53 mutations.
ChIP-seq analysis further elucidated the genomic landscape of ONECUT3 binding in MDS cells, revealing enrichment at key regulatory regions associated with chromosomal instability and mitotic defects. Notably, the identification of novel ONECUT3 transcriptional targets, including genes encoding components of the chromosomal passenger complex (CPC), provided mechanistic insights into ONECUT3-mediated pathogenesis in MDS.
Functional studies in cell culture models corroborated the oncogenic role of ONECUT3 in MDS, demonstrating that overexpression of ONECUT3 led to aberrant mitotic processes and increased chromosomal instability. Conversely, inhibition of ONECUT3’s transcriptional activity using a novel compound, C5484617, resulted in decreased cell proliferation and sensitization of MDS cells to treatment, highlighting the therapeutic potential of targeting ONECUT3 in high-risk MDS patients.
(J Clin Invest,2024 Feb 22:e172468.)
In conclusion, this study provides compelling evidence for the critical role of the transcription factor ONECUT3 in driving complex karyotypes and poor outcomes in Myelodysplastic Syndromes (MDS). Through a comprehensive analysis of patient samples and functional studies in cell culture models, we have demonstrated that elevated ONECUT3 expression levels are associated with specific genetic mutations and chromosomal instability in MDS, independently of TP53 status.
The identification of novel ONECUT3 transcriptional targets, including genes encoding components of the chromosomal passenger complex (CPC), provides mechanistic insights into ONECUT3-mediated pathogenesis and highlights potential therapeutic targets for intervention. Moreover, the discovery of a novel compound, C5484617, capable of inhibiting ONECUT3’s transcriptional activity and sensitizing MDS cells to treatment, represents a significant advancement in the development of targeted therapeutic strategies for high-risk MDS patients.
Moving forward, further validation studies and clinical trials are warranted to evaluate the efficacy and safety of targeting ONECUT3 in MDS patients. Additionally, efforts should be directed towards elucidating the molecular mechanisms underlying ONECUT3 dysregulation and exploring potential synergistic therapeutic approaches in combination with existing treatments.


