At the 2025 American Society of Hematology (ASH) Annual Meeting, Dr. Lucy Fox from the Peter MacCallum Cancer Centre (Australia) presented the highly anticipated results of the IBMDX study. This landmark trial evaluated the clinical utility and economic impact of "Upfront Whole Genome Sequencing (WGS)" in patients with suspected Inherited Bone Marrow Failure Syndromes (IBMFS). The findings suggest a paradigm shift is underway: WGS not only achieves a diagnostic yield of 37%—surpassing traditional panels and exome sequencing—but also uncovers complex genomic mechanisms previously invisible to standard testing.01. The Challenge: Unlocking the “Undiagnosed”
Inherited Bone Marrow Failure Syndromes represent a heterogeneous group of disorders that are notoriously difficult to diagnose due to overlapping clinical features and phenotypic variability. Traditional diagnostic strategies, such as targeted gene panels or Whole Exome Sequencing (WES), focus primarily on coding regions. Consequently, they often miss variants located in non-coding/regulatory regions or complex structural variants, leaving many patients without a definitive diagnosis.
To address this gap, the IBMDX study enrolled 237 pediatric and adult patients with suspected IBMFS. A key feature of the study design was the use of non-hematological DNA (primarily from hair bulbs) to rigorously distinguish germline variants from somatic mutations—a critical distinction in a patient population prone to hematologic malignancies.
02. Diagnostic Efficacy: WGS Breaks the Ceiling
Dr. Fox reported that upfront WGS achieved a diagnostic yield of 37%. While statistical modeling projected yields of 27% for targeted panels and 35% for WES, Dr. Fox emphasized that the numerical advantage of WGS extends far beyond a simple percentage increase. WGS provided a “one-stop” comprehensive analysis that identified:
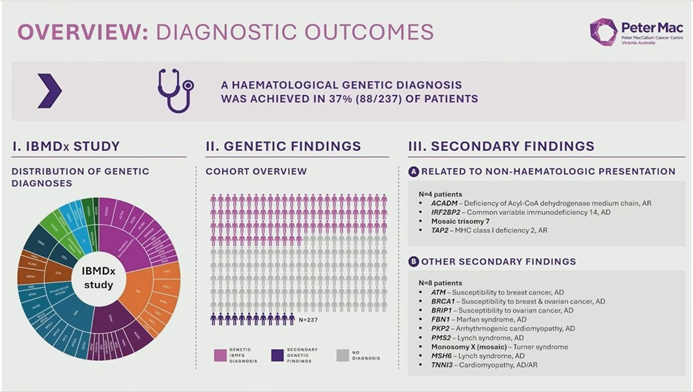
- Non-Hematological Diagnoses: Four patients received diagnoses unrelated to IBMFS that explained their symptoms (e.g., congenital hypothyroidism mimicking primary anemia).
- Secondary Findings: Clinically actionable secondary findings (e.g., cardiac or cancer risks) were identified in 8 patients, enabling preventive care.
03. Capturing the “Invisible”: Beyond the Exome
The true superiority of WGS was demonstrated through its ability to detect variants that are technically impossible for WES or panels to catch. Dr. Fox highlighted several specific mechanisms:
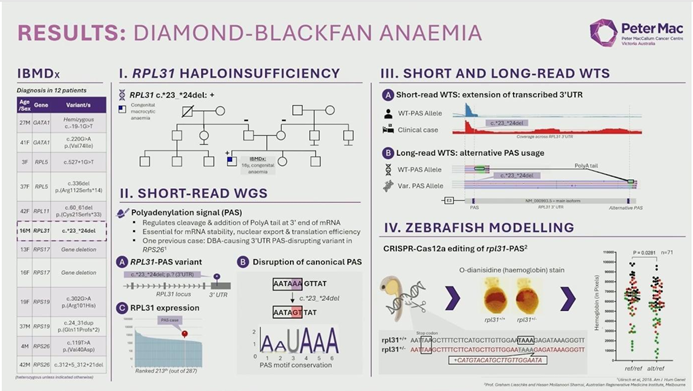
- Diamond-Blackfan Anemia (DBA): WGS identified a dinucleotide deletion in the 3′ UTR of the RPL31 gene. Because this variant lies in a non-coding region, it would be missed by WES. Functional validation confirmed this deletion disrupted a polyadenylation site, a mechanism further proven in zebrafish models.
- Severe Congenital Neutropenia (SCN): In a patient with a single heterozygous HAX1 mutation, WGS uncovered a “missing” second hit: a 3kb retrotransposon insertion within an intron. This complex structural variant is undetectable by standard short-read exome sequencing.
- Telomere Biology Disorders (TBD): WGS allowed for simultaneous variant detection and bioinformatic assessment of telomere length. A novel variant in the TERT pseudokinase domain was identified and functionally validated as pathogenic.
04. Economic Reality and Future Outlook
The study also applied a “Theoretical Framework of Acceptability” to assess feasibility.
- Cost vs. Benefit: Currently, the cost per diagnosis using upfront WGS is approximately $8,000 USD, compared to $4,600 USD for WES.
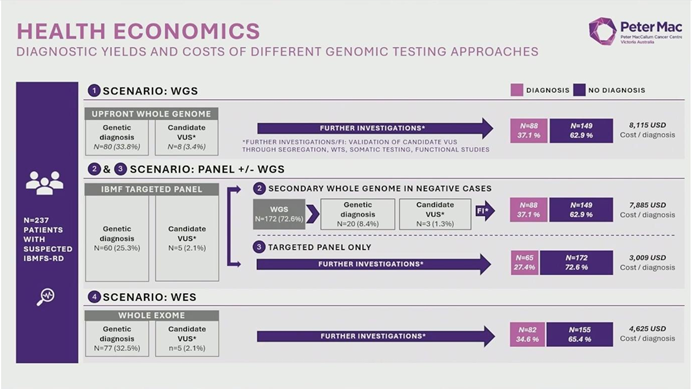
- The “Long-Tail” Value: Dr. Fox argued that the 37% yield is merely the “floor.” Unlike panels, WGS data can be re-analyzed indefinitely as new gene associations are discovered, increasing the yield over time without re-sequencing the patient.
Conclusion
Dr. Fox concluded that the IBMDX study validates Upfront Whole Genome Sequencing as a superior diagnostic tool for IBMFS. By integrating WGS with transcriptomics and functional validation, clinicians can now solve cases involving deep intronic variants, retrotransposons, and structural rearrangements that were previously diagnostic dead-ends.
While cost remains a short-term barrier, the inevitable decline in sequencing costs suggests WGS will soon become the standard of care. The massive dataset generated by IBMDX is now being housed in the global “Cardinal” platform, inviting international collaboration to further advance the understanding of these rare diseases.