In January 2023, a review by Professor Tao Cheng from Institute of Hematology & Blood Diseases Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College was published in the international academic journal ——Blood Science . The title of the study is "Spatially resolved transcriptomics: advances and applications". This review summarizes the interplay between technological innovation and biological discovery continues to push the boundaries of what was previously imaginable, translating our approach to complex biological systems and disease pathology.
Spatially resolved transcriptomics stands at the forefront of molecular biology and bioinformatics, merging the precision of genetic expression analysis with the complexity of spatial patterning. This confluence has been pivotal in dissecting the intricate tapestry of cellular and tissue architecture in health and disease.
Beyond smFISH (Single Molecule Fluorescence In Situ Hybridization) and seqFISH (Sequential Fluorescence In Situ Hybridization) , emerging techniques like hybridization chain reaction (HCR) FISH are enhancing multiplexing capabilities and sensitivity, enabling the visualization of a broader array of RNA molecules within their native tissue contexts. Advancements in microfluidics and nanotechnology have led to the development of more sophisticated sequencing-based approaches. Techniques such as microfluidic single-cell RNA sequencing (scRNA-seq) offer unprecedented throughput and sensitivity, facilitating the analysis of thousands of single cells in situ, providing a comprehensive view of cellular heterogeneity.
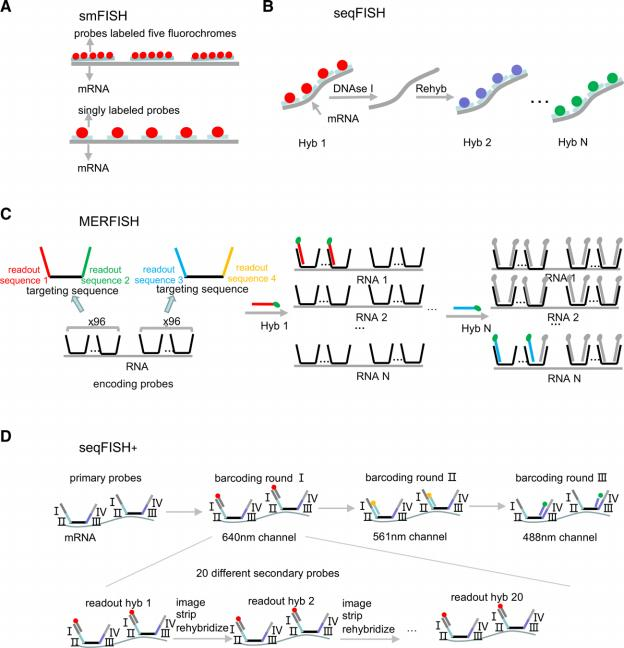
Figure,Schematics of the main imaging-based methods. (A) In smFISH, fluorochrome-labeled probes detect individual mRNA molecules through in situ hybridization. (B) In seqFISH, sequential barcoding: in each round of hybridization, a set of probes targeting different genes are hybridized on each transcript, imaged, and then stripped with DNAse I. The same probe sequences are used in different rounds of hybridization, but probes are coupled to different fluoro- phores. By decoding the temporal color sequences, the original genes can be identified. (C) In MERFISH, HD4 code is introduced by encoding probes contain- ing a central RNA-targeting region flanked by 2 readout sequences. Encoding probes convert the RNA into a unique combination of readout sequences. (D) In seqFISH+, I–IV sequences on the primary probes correspond to 4 rounds of barcoding, and the fourth round is used for error correction. Only one-twentieth of the total genes in each fluorescent channel are labeled by readout probes in each hybridization readout round, lowering the density of transcripts in each image. HD4 = minimum Hamming distance is 4, MERFISH = multiplexed error-robust FISH, seqFISH = seqFISH = sequential barcoded FISH, smFISH = sin- gle-molecule FISH.(Blood Science. 5(1):1-14, January 2023.)
Spatially resolved transcriptomics is revolutionizing our understanding of the spatial dimension of gene expression, offering a window into the complexity of biological systems. As we continue to push the frontiers of this technology, its integration with other omics technologies and computational tools will undoubtedly lead to groundbreaking discoveries in biology and medicine, offering new avenues for the diagnosis, treatment, and understanding of human diseases.
Reference:1. Duan, H; Cheng, T; Cheng, Hui. Spatially resolved transcriptomics: advances and applications. Blood Science. 5(1):1-14, January 2023.

